Quantitative analysis of endocrine disruption by ketoconazole and diethylstilbestrol in rat mammary gland development
Reproductive Toxicology
Below is a podcast and transcript of the interview concerning the 1st chapter of the book Bifurquer
Abstract
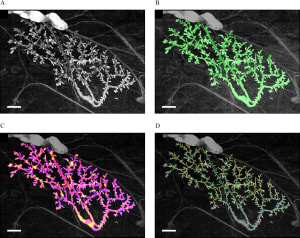
Endocrine disruptors alter mammary gland development, impair the ability to nourish offspring, and increase the cancer risk in animal models. Epidemiological studies reveal trends towards early mammary development, nursing problems, and breast cancer in younger women. Morphological changes in mouse postnatal mammary gland development are considered sensitive markers of endocrine disruption. While the mouse mammary gland is easily amenable to morphometric measurements from the fetal stage to full maturity, the rat mammary gland grows more conspicuously into the third dimension, hindering conventional morphometric analysis. However, since rats are more commonly used in international toxicological reproductive studies, it would be beneficial to include mammary gland whole-mount analysis in these studies. Using our quantitative software to perform computer-driven analysis of the rat mammary epithelium we examined the effects of gestational and postnatal exposure to ketoconazole, an antifungal medication that affects steroidogenesis, and to the estrogen diethylstilbestrol in the mammary glands of 6- and 22-day-old females. Both treatments produced effects at both ages; the epithelium was smaller and less complex in exposed animals compared to controls. Global analysis with the permutation test showed that morphological evaluation of the PND22 mammary gland is sensitive to endocrine disruption and possibly non-monotonic. In addition to revealing that ketoconazole altered the mammary gland structure, these results suggest that for future toxicology studies, day 22 (at weaning) is more suitable than day 6 because it showed significant measurements and trends. If the collection of mammary glands is added to existing international test methods, PND22 could be a relevant time point.
Keywords: Ketoconazole, Diethylstilbestrol, endocrine disruptors, perinatal exposure, mammary gland whole mount